Just as ChatGPT generates text by predicting the word most likely to follow in a sequence, a new artificial intelligence (AI) model can write new proteins that are not naturally ocurring from scratch.
Scientists used the new model, ESM3, to create a new fluorescent protein that shares only 58% of its sequence with naturally occurring fluorescent proteins, they said in a study published July 2 on the preprint bioRxiv database. Representatives from EvolutionaryScale, a company formed by former Meta researchers, also outlined details June 25 in a statement.
The research team has released a small version of the model under a non-commercial license and will make the large version of the model available to commercial researchers. According to EvolutionaryScale, the technology could be useful in fields ranging from drug discovery to designing new chemicals for plastic degradation.
ESM3 is a large language model (LLM) similar to OpenAI's GPT-4, which powers the ChatGPT chatbot, and the scientists trained their largest version on 2.78 billion proteins. For each protein, they extracted information about sequence (the order of the amino acid building blocks that make up the protein), structure (the three-dimensional folded shape of the protein), and function (what the protein does). They randomly masked pieces of information about these proteins and requested that ESM3 predict the missing pieces.
They scaled this model up from research that the same team was conducting while still at Meta. In 2022 they announced EMSFold — a precursor to ESM3 that predicted unknown microbial protein structures. That year, Alphabet's DeepMind also predicted protein structures for 200 million proteins.
Scientists subsequently pointed out that there are limitations to these AI models' predictions and that the protein predictions need to be verified. But the methods can still massively speed up the search for protein structures, because the alternative is to use X-rays to map out protein structures one by one — which is slow and costly.
ESM3 goes beyond just predicting existing proteins, however. Using the information gleaned from 771 billion unique pieces of information on structure, function and sequence, the model can generate new proteins with particular functions. It was described as a "ChatGPT moment for biology" by one of EvolutionaryScale's backers.
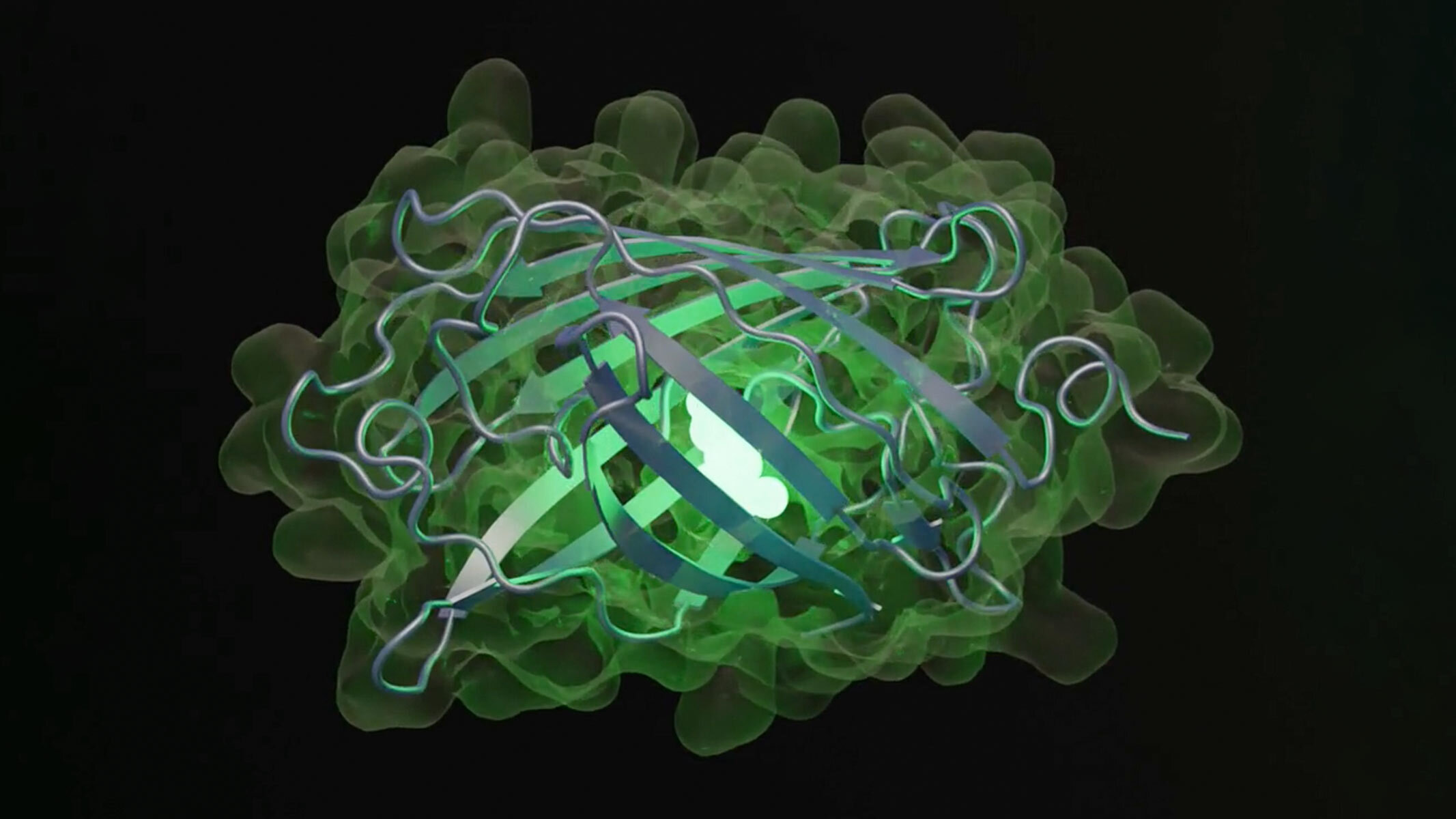
In the new study, the researchers queried the model to generate a new fluorescent protein — a kind of protein that captures light and releases it back at a longer wavelength, making it shine in a new shade of green. These proteins are important for biological researchers who append them to molecules that they're interested in studying to track and image them; their discovery and development won a Nobel Prize in chemistry in 2008.
The model generated 96 proteins with sequences and structures likely to produce fluorescence. The researchers then chose one with the fewest sequences in common with naturally fluorescent proteins. Although this protein was 50 times less bright than natural green fluorescent proteins, ESM3 generated another iteration that led to new sequences that increased brightness — and the result was a green fluorescent protein unlike any found in nature, dubbed "esmGPF." These iterations, done in moments by the AI, would take 500 million years of evolution to achieve, the EvolutionaryScale team estimated.







