The recent outbreak of monkeypox virus has called into question the capabilities of these kinds large DNA viruses to evolve, adapt and change their biology.
Compared with small RNA viruses such as coronavirus, monkeypox virus and other large DNA viruses are thought to evolve slowly. Yet there’s clear evidence that this really isn’t a hindrance to these viruses. In fact, they can adapt to new environments like us.
Although most infections remain mild, monkeypox can be a serious life-threatening disease, resulting in sepsis, encephalitis (brain inflammation) and blindness. The most common symptoms are rash and skin lesions, alongside flu-like symptoms and swollen lymph nodes.
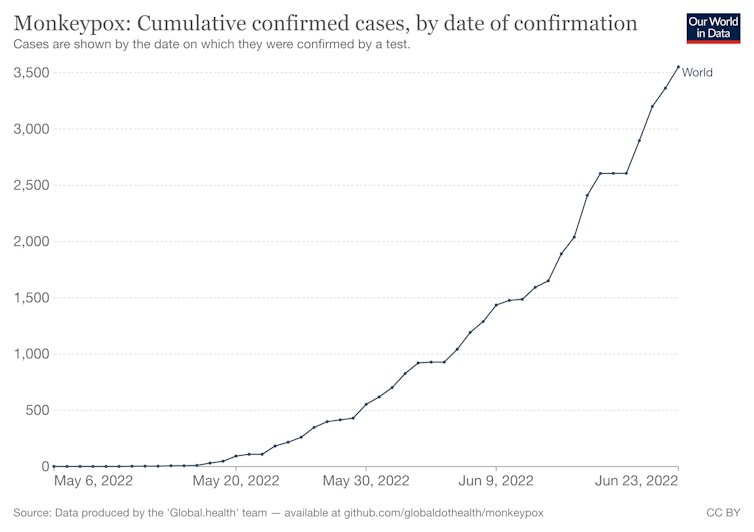
Cumulative monkeypox cases in current outbreak
Monkeypox virus naturally infects wild rodents, such as squirrels and rats, in west and central Africa – but it can jump species into humans and other animals. However, once it has jumped to humans, it cannot keep transmission going and eventually outbreaks die out. This is probably because monkeypox has not adapted itself to its new environment of humans, as spillback into wild rodents from infected humans is unlikely.
Monkeypox is closely related to the viruses that caused smallpox (variola viruses) and the virus that we use to vaccinate and eradicate smallpox (vaccinia virus). This group of viruses, referred to as poxviruses, are a kind of large DNA virus, meaning that their genome is composed of a chemical known as DNA, like our genome. (Coronavirus and related viruses use a cousin molecule called RNA.)
Other DNA viruses are the large DNA viruses adenoviruses and herpesviruses, but also small ones like papillomaviruses and parvoviruses. The viral genomes composed of either DNA or RNA essentially are the instructions to make new viruses, infect us and cause disease. Changes to the instructions can change virus biology.
As we have seen with SARS-CoV-2 and its variants, viruses can change how they behave with regards to spreading, disease severity and vaccine sensitivity. This is because of changes accumulating in the virus genome. Virus replication generates diversity in its genome, which can be acted on by evolutionary forces such as natural selection, to increase in frequency and maybe even out-compete older versions.
Evolutionary changes can occur when the virus encounters a new environment that it is not fully adapted to. Although all viruses can evolve rapidly due to their vast population sizes and rapid generation times, RNA viruses are thought to be masters of evolution because they have high mutation rates due to their small size, and many often lack error-correcting ability meaning more mutations occur every time they replicate.
Poxviruses have some characteristics that make them more generalist, including stable infectious particles, giving them more chances to infect. They use very common molecules on your cells to gain entry and infect, unlike SARS-CoV-2 which needs the specific ACE2 protein to gain entry to our cells.
Large DNA viruses such as monkeypox also contain lots of genes that target and manipulate different parts of the immune system.
Room for improvement
However, there’s clear evidence that improvements can be made, because, in humans, monkeypox transmission is relatively inefficient, with long incubation periods.
In general, large DNA viruses such as monkeypox are no different from other viruses, and their mutability is the basis for our ability to track and trace monkeypox outbreaks. They make mistakes and errors accumulate, which can be used as fuel for evolution and biological changes. There’s even evidence from the recent monkeypox outbreak that the host cell is directly mutating the virus genome.
Studies focusing on related poxviruses like the vaccinia virus have even uncovered new tricks they can use, which include rapidly amplifying the number of genes they use to attack our immune system. They could even borrow some of our own genes to help them infect us.
We can’t predict the trajectory that monkeypox evolution will take, so we must take the threat of this virus adapting to its new hosts (humans) seriously. And we need to use all the public health tools at our disposal to halt the current outbreak in all countries – including those where it is endemic.
Connor Bamford receives funding from UKRI, Wellcome Trust, British Medical Association Foundation, SFI.
This article was originally published on The Conversation. Read the original article.