The Earth Biogenome Project, a global consortium that aims to sequence the genomes of all complex life on earth (some 1.8 million described species) in ten years, is ramping up.
The project’s origins, aims and progress are detailed in two multi-authored papers published today. Once complete, it will forever change the way biological research is done.
Specifically, researchers will no longer be limited to a few “model species” and will be able to mine the DNA sequence database of any organism that shows interesting characteristics. This new information will help us understand how complex life evolved, how it functions, and how biodiversity can be protected.
The project was first proposed in 2016, and I was privileged to speak at its launch in London in 2018. It is currently in the process of moving from its startup phase to full-scale production.
The aim of phase one is to sequence one genome from every taxonomic family on earth, some 9,400 of them. By the end of 2022, one-third of these species should be done. Phase two will see the sequencing of a representative from all 180,000 genera, and phase three will mark the completion of all the species.
The importance of weird species
The grand aim of the Earth Biogenome Project is to sequence the genomes of all 1.8 million described species of complex life on Earth. This includes all plants, animals, fungi, and single-celled organisms with true nuclei (that is, all “eukaryotes”).
While model organisms like mice, rock cress, fruit flies and nematodes have been tremendously important in our understanding of gene functions, it’s a huge advantage to be able to study other species that may work a bit differently.
Many important biological principles came from studying obscure organisms. For instance, genes were famously discovered by Gregor Mendel in peas, and the rules that govern them were discovered in red bread mould.
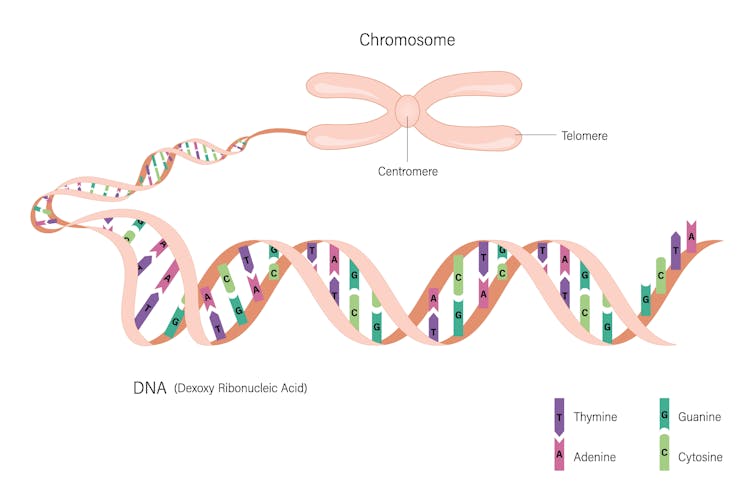
DNA was discovered first in salmon sperm, and our knowledge of some systems that keep it secure came from research on tardigrades. Chromosomes were first seen in mealworms and sex chromosomes in a beetle (sex chromosome action and evolution has also been explored in fish and platypus). And telomeres, which cap the ends of chromosomes, were discovered in pond scum.
Answering biological questions and protecting biodiversity
Comparing closely and distantly related species provides tremendous power to discover what genes do and how they are regulated. For instance, in another PNAS paper, coincidentally also published today, my University of Canberra colleagues and I discovered Australian dragon lizards regulate sex by the chromosome neighbourhood of a sex gene, rather than the DNA sequence itself.
Read more: Sex lives of reptiles could leave them vulnerable to climate change
Scientists also use species comparisons to trace genes and regulatory systems back to their evolutionary origins, which can reveal astonishing conservation of gene function across nearly a billion years. For instance, the same genes are involved in retinal development in humans and in fruit fly photoreceptors. And the BRCA1 gene that is mutated in breast cancer is responsible for repairing DNA breaks in plants and animals.
The genome of animals is also far more conserved than has been supposed. For instance, several colleagues and I recently demonstrated that animal chromosomes are 684 million years old.
Read more: Specks of dust on the microscope slide? No, we are looking at the building blocks of our genome
It will be exciting, too, to explore the “dark matter” of the genome, and reveal how DNA sequences that don’t encode proteins can still play a role in genome function and evolution.
Another important aim of the Earth Biogenome Project is conservation genomics. This field uses DNA sequencing to identify threatened species, which includes about 28% of the world’s complex organisms – helping us monitor their genetic health and advise on management.
No longer an impossible task
Until recently, sequencing large genomes took years and many millions of dollars. But there have been tremendous technical advances that now make it possible to sequence and assemble large genomes for a few thousand dollars. The entire Earth Biogenome Project will cost less in today’s dollars than the human genome project, which was worth about US$3 billion in total.
In the past, researchers would have to identify the order of the four bases chemically on millions of tiny DNA fragments, then paste the entire sequence together again. Today they can register different bases based on their physical properties, or by binding each of the four bases to a different dye. New sequencing methods can scan long molecules of DNA that are tethered in tiny tubes, or squeezed through tiny holes in a membrane.
Why sequence everything?
But why not save time and money by sequencing just key representative species?
Well, the whole point of the Earth Biogenome Project is to exploit the variation between species to make comparisons, and also to capture remarkable innovations in outliers.
There is also the fear of missing out. For instance, if we sequence only 69,999 of the 70,000 species of nematode, we might miss the one that could divulge the secrets of how nematodes can cause diseases in animals and plants.
There are currently 44 affiliated institutions in 22 countries working on the Earth Biogenome Project. There are also 49 affiliated projects, including enormous projects such as the California Conservation Genomics Project, the Bird 10,000 Genomes Project and UK’s Darwin Tree of Life Project, as well as many projects on particular groups such as bats and butterflies.
Jenny Graves is a member of the Earth Biogenome Project Working Group, and an author on the two PNAS papers. She receives funding for other projects from the Australian Research Council.
This article was originally published on The Conversation. Read the original article.







